Look discovers mobile reveal that hints recycling is in our DNA – Google

This article has been reviewed primarily based entirely mostly on Science X’s editorial course of and insurance policies. Editors possess highlighted the next attributes while guaranteeing the shriek’s credibility:
reality-checked
stare-reviewed publication
relied on source
proofread
by Rose Miyatsu, University of California – Santa Cruz
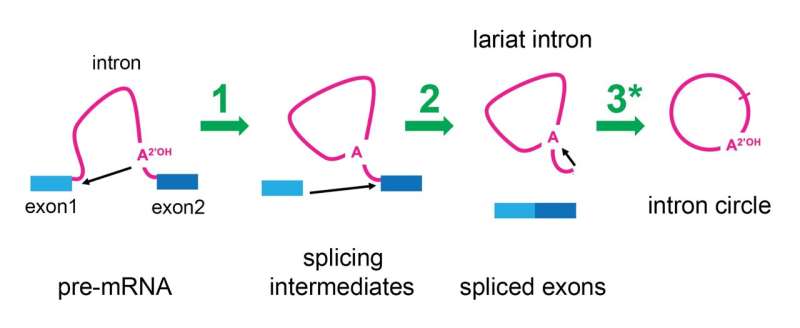
Although you must maybe no longer enjoy them, or possess even heard of them, for the length of your body, limitless little machines known as spliceosomes are laborious at work. As you sit down and read, they are faithfully and all straight away inserting lend a hand together the broken info to your genes by attempting down sequences known as “introns” so that your messenger RNAs can accomplish the suitable proteins wished by your cells.
Introns are maybe one of our genome’s very most keen mysteries. They are DNA sequences that interrupt the excellent protein-coding info to your genes, and want to be “spliced out.” The human genome has heaps of of thousands of introns, about 7 or 8 per gene, and every is removed by a in actuality educated RNA protein complicated known as the “spliceosome” that cuts out the entire introns and splices together the final coding sequences, known as exons. How this methodology of broken genes and the spliceosome developed in our genomes is no longer known.
Over his prolonged occupation, Manny Ares, UC Santa Cruz accepted professor of molecular, mobile, and developmental biology, has made it his mission to be taught as mighty about RNA splicing as he can.
“I’m all in regards to the spliceosome,” Ares stated. “I trusty want to hold the entirety the spliceosome does—even if I accomplish no longer know why it is doing it.”
In a new paper printed within the journal Genes and Pattern, Ares reports on an very excellent attempting discovery in regards to the spliceosome that will sing us more in regards to the evolution of varied species and the advance cells possess tailored to the uncommon challenge of introns. The authors point to that after the spliceosome is performed splicing the mRNA, it stays crammed with life and can have interaction in extra reactions with the removed introns.
This discovery presents the strongest indication we now possess up to now that spliceosomes would maybe be in a position to reinsert an intron lend a hand into the genome in a single other arrangement. Here’s an capability that spliceosomes weren’t beforehand believed to possess, however which is a general characteristic of “Team II introns,” distant cousins of the spliceosome that exist primarily in bacteria.
The spliceosome and Team II introns are believed to fragment a general ancestor that used to be accountable for spreading introns for the length of the genome, however while Team II introns can splice themselves out of RNA and then straight lend a hand into DNA, the “spliceosomal introns” that are shroud in most higher-level organisms require the spliceosome for splicing and weren’t believed to be reinserted lend a hand into DNA. On the opposite hand, Ares’s lab’s finding signifies that the spliceosome could maybe well aloof be reinserting introns into the genome at the present time. Here’s an arresting likelihood to place in thoughts because introns that are reintroduced into DNA add complexity to the genome, and dealing out more in regards to the establish these introns approach from could maybe well support us to better fee how organisms proceed to adapt.
Constructing on a spell binding discovery
An organism’s genes are comprised of DNA, whereby four bases, adenine (A), cytosine (C), guanine (G) and thymine (T) are ordered in sequences that code for organic instructions, esteem how to accomplish explicit proteins the body wants. Sooner than these instructions could maybe well furthermore be read, the DNA gets copied into RNA by a course of in most cases known as transcription, and then the introns in that RNA want to be removed sooner than a ribosome can translate it into trusty proteins.
The spliceosome gets rid of introns the employ of a two-step course of that finally ends up within the intron RNA having one of its ends joined to its center, forming a circle with a tail that appears to be like esteem a cowboy’s “lariat,” or lasso. This look has led to them being named “lariat introns.” No longer too prolonged within the past, researchers at Brown University who were studying the locations of the joining sites in these lariats made an uncommon observation—some introns were in actuality round as a alternative of lariat fashioned.
This observation straight bought Ares’s attention. Something looked as if it would be interacting with the lariat introns after they were removed from the RNA sequence to alternate their form, and the spliceosome used to be his main suspect.
“I belief that used to be keen attributable to this used, used thought in regards to the establish introns got here from,” Ares stated. “There could be a good deal of proof that the RNA parts of the spliceosome, the snRNAs, are carefully associated to Team II introns.”
Since the chemical mechanism for splicing is incredibly identical between the spliceosomes and their distant cousins, the Team II introns, many researchers possess theorized that after the course of of self-splicing turned into too inefficient for Team II introns to reliably entire on their very possess, parts of these introns developed to become the spliceosome. While Team II introns were in a position to insert themselves straight lend a hand into DNA, on the opposite hand, spliceosomal introns that required the support of spliceosomes weren’t belief to be inserted lend a hand into DNA.
“One in all the questions that used to be vogue of missing from this epic in my thoughts used to be, is it doable that the in vogue spliceosome is aloof in a position to rob a lariat intron and insert it someplace within the genome?” Ares stated. “Is it aloof in a position to doing what the ancestor complicated did?”
To launch as a lot as acknowledge to this quiz, Ares decided to analyze whether it used to be certainly the spliceosome that used to be making adjustments to the lariat introns to rob away their tails. His lab slowed the splicing course of in yeast cells, and positioned that after the spliceosome launched the mRNA that it had performed splicing introns from, it hung onto intron lariats and reshaped them into dazzling circles. The Ares lab used to be in a position to reanalyze printed RNA sequencing info from human cells and positioned that human spliceosomes also had this capability.
“We’re smitten by this because while we accomplish no longer know what this round RNA could maybe well originate, the truth that the spliceosome is aloof crammed with life suggests it would maybe be in a position to catalyze the insertion of the lariat intron lend a hand into the genome,” Ares stated.
If the spliceosome is willing to reinsert the intron into DNA, this would maybe also add principal weight to the speculation that spliceosomes and Team II introns shared a general ancestor prolonged within the past.
Checking out a principle
Now that Ares and his lab possess proven that the spliceosome has the catalytic capability to hypothetically space introns lend a hand into DNA esteem their ancestors did, the next step is for the researchers to possess an synthetic scenario whereby they “feed” a DNA strand to a spliceosome that is aloof linked to a lariat intron and glimpse if they are able to in actuality web it to insert the intron someplace, which would shroud “proof of thought” for this principle.
If the spliceosome is willing to reinsert introns into the genome, it is more likely to be a truly uncommon occasion in humans, because the human spliceosomes are in incredibly high request and therefore originate no longer possess mighty time to employ with removed introns. In assorted organisms the establish the spliceosome is no longer in actuality as busy, on the opposite hand, the reinsertion of introns would maybe be more frequent. Ares is working carefully with UCSC Biomolecular Engineering Professor Russ Corbett-Detig, who has lately led a scientific and exhaustive hunt for new introns within the on hand genomes of all intron-containing species that used to be printed within the journal Complaints of the Nationwide Academy of Sciences (PNAS) final yr.
The paper in PNAS confirmed that intron “burst” occasions a long way lend a hand in evolutionary history likely provided thousands of introns into a genome all straight away. Ares and Corbett-Detig are in actuality working to recreate a burst occasion artificially, which would give them insight into how genomes reacted when this took space.
Ares stated that his corrupt-disciplinary partnership with Corbett-Detig has opened the doorways for them to in actuality dig into about a of the very most keen mysteries about introns that it would maybe be no longer doable for them to price fully without their combined skills.
“It is truly the most easy advance to originate things,” Ares stated. “Must you witness any individual who has the same roughly questions in thoughts however a sure space of suggestions, perspectives, biases, and peculiar suggestions, that gets more thrilling. That makes you’re feeling esteem you would maybe web away and solve a challenge esteem this, which is incredibly complicated.”
More info: Manuel Ares et al, Intron lariat spliceosomes convert lariats to dazzling circles: implications for intron transposition, Genes & Pattern (2024). DOI: 10.1101/gad.351764.124
Citation: Look discovers mobile reveal that hints recycling is in our DNA (2024, May well well well furthermore 11) retrieved 11 May well well well furthermore 2024 from https://phys.org/news/2024-05-mobile-hints-recycling-dna.html
This story is field to copyright. Other than any dazzling dealing for the explanation for non-public look or research, no share would maybe be reproduced without the written permission. The shriek is geared up for info functions most efficient.

